Design
PFIO is an IO abstraction library for Chainer, optimized for deep learning training with batteries included. It supports
Filesystem API abstraction with unified error semantics,
Explicit user-land caching system,
IO performance tracing and metrics stats, and
Fileset container utilities to save metadata.
Rationale
There are a lot of non-POSIX-compliant filesystems in the industry and all of them have Pros and Cons, from cloud storages like Google Cloud Storage and Amazon S3, to on-premise distributed filesystems like HDFS. Supporting different filesystems by developers themselves creates unnecessary burden on the developers and might reduce the portability on the code. That is the motivation of PFIO supporting filesystem abstraction API.
Also, deep learning training programs have benefited from filesystem page caching provided Linux kernel because its optmization method is based on stochastic gradient descent, where all training data is repeatedly read to iteratively train the model. But such non-POSIX-compliant filesystem usually does not provide content caching capability and I/O workload of training program would not implicitly optmized unlike Posix-based filesystems in Linux. To help with learning on non-POSIX-compliant filesystems, e.g. HDFS, PFIO implements data caching capability in userland. Moreover, developers can choose which data to cache, from the raw files to DNN input NumPy array.
Opmization in deep learning model training is also important as the model training usually takes long time and even 1% speedup is important. Modern external non-Posix filesystem is based on complex communication protocol between multiple data nodes and its performance metrics are not simply observable. Built-in tracing system, would be the first step for optimization, mitigating the complexity problem.
One of the general problems in filesystems is scalability of metadata access, caused by inbalanced ratio of number of files and total capacity of one system. PFIO supports various container file formats to aggregate many small files into single large file with metadata mapping, e.g. HDF5, ZIP and Tar (and more in future), by taking advantage of the fact that in machine learning training workload, usually SGD, is repeated read of single fixed dataset (Write-once-and-read-many). Aggregating millions of single-kilobytes files into single file would save a lot of metadata store.
Architecture
PFIO abstracts underlying system with three objects. See API documentation for details.
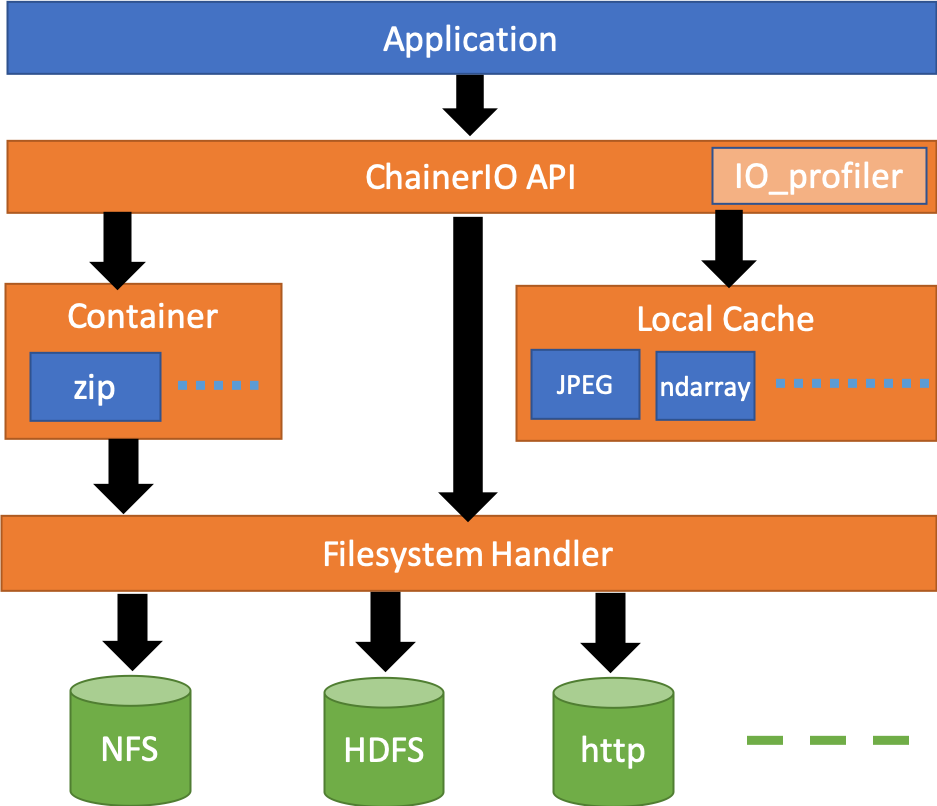
FileSystem
Abstraction of each filesystems. Depending on the context the term might stand for the filesystem type, or the (network) filesystem instance. It supports
Getting basic information of the filesystem (info)
Container creation, deletion
Accessing containers
Accessing raw files
Listing all files under specific directory
Primarily HDFS and POSIX, and AWS S3 API
FS
Abstraction of a directory subtree, or file containers such as ZIP. It contains a set of (key, binary object) pairs. Keys are typically path-like string and binary is typically a file content. In PFIO keys in a container are UTF-8 strings. Containers can be nested, e.g. ZIP in ZIP. It supports:
Showing basic information of the container (info)
Accessing raw files included (open)
Accessing containers included (open)
Adding and remove file (create, delete)
Listing keys in a container
Primarily ZIP, and possibly Hdf5?
import pfio
from PIL import Image
import io
with pfio.v2.from_url('hdfs://name-service-cluster1/some/many-files-dataset.zip') as fs:
print(container.info())
# List all keys in the container
for name in fs.list(recursive=True):
print(name)
# Obtains a file object to access binary content that
# corresponds to the key ``some/file.jpg``
with fs.open('some/file.jpg', 'rb') as fp:
binary = fp.read()
image = Image(io.BytesIO(binary))
...
Custom Scheme
Note
This is an experimental feature.
The format of URL is defined by the URL standard in WhatWG . Developers and users can configure
a custom scheme with a predefined scheme handler. The primary use case
is a private cloud environment that has an S3-compatible object
storage. Reaching out with custom endpoint is supported by
pfio.v2.S3, but custom endpoints are only configurable
with keyword arguments. With custom schemes, arbitrary configurations
are injectable with configuration file and they’re available with
custom scheme name. For example, a config file like:
[foobar]
scheme = s3
endpoint = https://foobar.example.com/
aws access_key_id = $CUSTOM_ACCESS_KEY
aws_secret_access_key = $CUSTOM_ACCESS_SECRET
will be available like this in Python:
fs = pfio.v2.from_url('foobar://path/to/a')
with fs.open('filename') as fp:
fp.read()
It means you can open an object named /path/to/a/filename in an
S3-compatible system with an endpoint foobar.example.com, with
pfio.v2.S3 and with the specified access key. Technically,
except the scheme property, they are keyword arguments for
pfio.v2.from_url() . The above example code is equal to:
key_id = os.getenv("CUSTOM_ACCESS_KEY")
secret = os.getenv("CUSTOM_ACCESS_SECRET")
fs = pfio.v2.from_url('s3://path/to/a',
endpoint="https://foobar.example.com",
aws_access_key_id=key_id,
aws_secret_access_key=secret)
with fs.open('filename') as fp:
fp.read()
PFIO reads $PFIO_CONFIG_PATH for the first hand. If the
environment variable is undefined, it tries to open and load located
as $XDG_CONFIG_HOME/pfio.ini.
See also:
File-like Objects
Abstraction of binary objects or files, typically returned by
fs.open method. It is an implementation of RawIOBase class
(See RawIOBase in Python
document). It supports
Read to underlying file or binary in a container
Writes supported by filesystems, but possibly not in containers
URI Expression of File Paths
Filesystems can be expressed as:
<filesystem> := <scheme>[://<service-idenfier>]
where scheme represends the filesystem type. Currently hdfs
and file are supported. hdfs stands for HDFS and file
means local filesystem. For remote network file system like HDFS,
service-identifier stands for service instance. It can be omitted
when the default service is defined. For example in HDFS, it is the
name of name service described in hdfs-site.xml in the Hadoop
configuration directory like following example:
<configuration>
<property>
<name>dfs.nameservices</name>
<value>hdfs-nameservice1</value>
</property>
...
In this example is service-identifier is hdfs-nameservice.
Containers, files are uniquely identifiable by partial set of URI expression:
<uri> := <scheme>://[<service-idenfier>]/<path>
service-identifier can be omitted when it can be uniquely defined
by the environment. path is a UTF-8 string, a sequence of path
segments separated by / and path segments are recommended to only
use [a-z][A-Z][0-9][_-] . However, details depend on underlying filesystem
implementation or containers.
pfio.open_as_container and pfio.fs.open take
filesystem, uri or path as an argument to identify the
file to be opened, when the context is a filesystem. If the context
is a container, they accept a key as an argument.
If the context is a file system, they also take a path as a
relative path. The base for relative path depends on filesystems; for
HDFS it is home directory and for POSIX it is current working
directory.
For example, all these fs.open open the same file, given that the
default name service is name-service1 and user Smith’s home
derectory is defined as /user/smith:
import pfio
# Using full URI
pfio.open('hdfs://name-service1/user/smith/path/to/file.txt')
# Using set_root and absolute path
pfio.set_root('hdfs://name-service1/')
pfio.open('/user/smith/path/to/file.txt')
# Using set root and relative path
pfio.set_root('hdfs')
pfio.open('path/to/file.txt')
# Overwrite the global setting with full URI
# Access the posix with the global setting to hdfs
pfio.open('file://path/to/file.txt')
# Accessing with filesystem object
with pfio.create_handler('hdfs') as handler:
handler.open('file.txt')
Major Use Cases
With all these primitive concepts and operations PFIO supports various use cases from loading training data, taking snapshots of models in the middle of training process, and recording the final model.
In order to load training data in Chainer, developers create a dataset class which derived the DatasetMixin from the chainer.dataset package. PFIO will provide several implementation replacements for generic datasets included in Chainer and other Chainer family libraries.
According to the survey we conduct on developers’ code. I/Os can be categorized into two different classes.
Inputs and outputs using file object: direct access via built-in APIs e.g. Image class in PIL, cv2.image.open and pandas.read_hdf. In such case, the file object (in PFIO, it is implementation of RawIOBase )
Inputs and outputs all wrapped by 3rd party library. Some of them has functions only takes the file path string as an argument and all file operations are hidden underneath the library. Examples are cv2.VideoWriter(), cv2.imread() and cv2.VideoCapture() from OpenCV. Since we cannot change the library, we provide a monkey patch of major libraries frequently used along with Chainer.
For details see API.
V2 API history
PFIO v2 API tries to solve the impedance mismatch between different local filesystem, NFS, and other object storage systems, with a lot simpler and cleaner code.
It has removed several toplevel functions that seem to be less
important. It turned out that they introduced more complexity than
originally intended, due to the need of the global context. Thus,
functions that depends on the global context such as open(),
set_root() and etc. have been removed in v2 API.
Instead, v2 API provides only two toplevel functions that enable
direct resource access with full URL: open_url() and
from_url(). The former opens a file and returns FileObject. The
latter, creates a fs.FS object that enable resource access under
the URL. The new class fs.FS, is something close to handler object
in version 1 API. fs.FS is intended to be as much compatible as
possible, however, it has several differences.
One notable difference is that it has the virtual concept of current
working directory, and thus provides subfs() method. subfs()
method behaves like chroot(1) or os.chdir() without actually
changing current working directory of the process, but actually
returns a new fs.FS object that has different working
directory. All resouce access through the object automatically
prepends the working directory.
V2 API does not provide lazy resouce initialization any more. Instead,
it provides simple wrapper lazify(), which recreates the fs.FS
object every time the object experiences fork(2). Hdfs and
Zip can be wrapped with it, and will be fork-tolerant object.